| |
It’s perhaps the biggest question in biology: What is life?
On the most microscopic level, life begins with sequences of genetic information that encode
proteins,
which then form the molecular machinery of individual cells. Groups of cells working together can form all sorts of living organisms from bacteria to plants, animals and humans. There are only a small number of
genes
and proteins that can actually sustain life, which may lead us to believe that they are somehow special or unique. But is this actually true?
A new field of biology, known as
synthetic biology,
is testing the boundaries of what constitutes life by combining biological, chemical and engineering techniques to try to alter living systems in nature and perhaps even develop new ones. Recently, Princeton chemist Dr. Michael Hecht has shown that a strain of the bacteria
Escherichia coli
can survive using genetic sequences created in the laboratory. These sequences are found nowhere else in nature.
“We started off with a basic question,” Dr. Hecht explains. “Is it possible to sustain the growth of cells using genes and proteins constructed from scratch in the laboratory?”
Building the Novel Protein
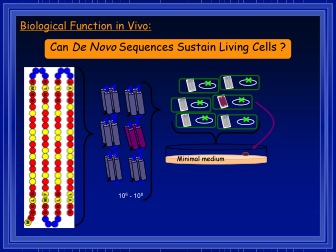
Biological function in vivo. Courtesy Dr. Michael Hecht.
To find out, Dr. Hecht first had to create the novel proteins from artificial sequences of
DNA.
This is not a simple process. Genetic sequences are composed of varying arrangements of four
nucleotides:
adenine, guanine, thymine and cytosine, strung together to create strands of DNA. Specific sets of three nucleotides indicate particular
amino acids.
For example, three adenines together encode the amino acid lysine, whereas two adenines followed by a cytosine encode a different amino acid, asparagine. There are a total of 20 amino acids that, when strung together in a chain, form the underlying structure of all proteins. Through a process known as
translation,
hundreds to thousands of pairs of nucleotides provide the instructions for constructing the sequences of each individual protein.
However, not all sequences of amino acids can form functional proteins. To become a functional protein, the amino acid sequence of a protein must first fold into a specific 3-dimensional structure. This folding is mediated by contacts between the amino acids in the chain. Since each of the 20 amino acids has a different shape, charge and “fear” of water (or hydrophobicity), some will attract one another, while others will repel. Additionally, some will be attracted to water on the outer surface of the protein, while others will avoid water by becoming buried in the interior of the protein. Ultimately the folding of each protein into its unique 3-dimensional structure depends on the ordering of the amino acids in its sequence. [See What A Year! for November, 2006, on protein folding and Parkinson’s disease.]
The construction of viable folded proteins, then, could not begin from random collections of DNA sequences. Rather, Dr. Hecht and his students developed a strategy that arranged amino acids into a sequence based on their charge (polarity) and hydrophobicity, thereby ensuring that they would fold correctly. They then worked backwards from the arrangement of amino acids to put together the genetic sequence that would encode for the desired protein. Over the course of several years, Dr. Hecht and his team of researchers created 2 million proteins never before seen in nature.
E. coli
The next step was to figure out whether a living organism could actually use one of these artificial proteins to survive. Dr. Hecht’s group used E. coli bacteria as a model living organism. Normal bacterial cells can synthesize all their essential amino acids and can therefore grow on what is called a minimal medium, which consists of just sugars and salts. But if the bacterium is missing a gene involved in the synthesis of an amino acid, it will not be able to grow on minimal medium.
Dr. Hecht and his students tested 27 E. coli strains that each lacked a different essential gene. This means that none of them would grow on minimal medium, although they would grow on a medium that provided them with full nutrition. In each experiment, billions of cells from one of the 27 E. coli strains were mixed together in a test tube with genes encoding the 2 million artificial proteins that Dr. Hecht had created in his laboratory. The cells were then shocked with an electric impulse to make them take up the DNA encoding the novel proteins. The cells were then placed on minimal medium and Dr. Hecht and his students observed their growth. Any cell that didn’t take up DNA or that took up a piece of DNA that didn’t rescue the deleted function would not grow. On the rare occasion that a cell took up a piece of artificial DNA that did rescue the function that had been deleted in its genome, it would grow to be a visible colony about the size of a pinhead. This was a really exciting result because it showed that growth of living cells could be sustained by an artificial protein not found in nature, which answered Dr. Hecht’s initial question.
At this point in the experiment, Dr. Hecht and his students still didn’t know which of the 2 million artificial proteins had been the one to “rescue” the defective cells and allow them to grow. To find out, they took samples of cells from the colony and sequenced the relevant piece of DNA to determine the sequence of nucleotides.
The experiment was then repeated for each of the 27 strains of E. coli. In many cases, there was no growth, indicating that no artificial sequences in the collection could rescue the deletion in E. coli. However in several cases, colonies indeed grew, demonstrating that a novel sequence, which had never before existed in nature, could sustain the growth of living cells. Ultimately, they were able to identify four deletions that could be rescued by artificial sequences that did not exist in nature, but had been created in the lab.
Next they tested whether all four artificial sequences could work simultaneously by introducing all four into E. coli cells that lacked all four genes. The experiment worked. The multiply defective cells were rescued by the four novel sequences.
Some Arithmetic
To put this result into a bit of context, the E. coli genome contains about 4,000 genes. Replacing four of these 4,000 genes means replacing 0.1% of the genome with artificial proteins. Of these 4,000 genes, only about 400 are essential for growth on minimal media, which means 4/400 or 1% of the most essential genes in the genome have been replaced by artificial proteins. That is quite amazing.
Next Steps
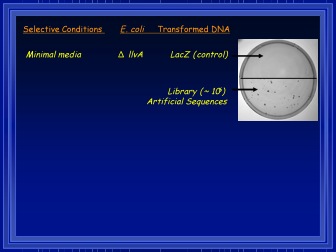
Rescued vs. control. Courtesy Dr. Michael Hecht.
Now Dr. Hecht and his research group are moving forward in two directions: First, they are trying to determine exactly how the artificial proteins rescue the deletions. At first glance, it may seem that a deletion must be rescued by a protein providing the same function as the deleted natural protein. However, this is not necessarily the case. Life is opportunistic, and there can be more than one way to achieve a particular goal. Thus, the artificial proteins may sustain life by mechanisms that are significantly different from those used by naturally evolved proteins.
The second main goal in Hecht’s lab is to see how many genes can be replaced simultaneously. Would it be possible, for example, to replace 10% of the E. coli genome with artificial proteins? If so, would it still be called E. coli or would it be a new form of life? Dr. Hecht and his students have also begun similar experiments in yeast,
eukaryotic
organisms that are more highly evolved cells. Dr. Hecht’s results and his ongoing research continue to push the boundaries of life as we know it.
Dr. Michael Hecht is a Professor of Chemistry at Princeton
University. He is also the Master of Forbes
College, one of the six residential colleges at Princeton. His research focuses on the creation of artificial genes and proteins not found in nature, which have the capability to perform a variety of biological functions. When not teaching or in the laboratory, Dr. Hecht enjoys hiking, skiing, and spending time with his family.
For More Information:
- Fisher, M. et al. 2011. “De Novo Designed Proteins from a Library of Artificial Sequences Funtion in Escherichia Coli and Enable Cell Growth.” PLoS One, 6(1).
- Patel, S. et al. 2009. “Cofactor binding and enzymatic activity in an unevolved superfamily of de novo designed 4-helix bundle proteins.” Protein Science, 18: 1388-1400.
- Hecht, M. et al. 2004. “De novo proteins from designed combinatorial libraries.” Protein Science, 13: 1711-1723.
- Kamtekar, S. et al. 1993. “Protein Design by Binary Patterning of Polar and Nonpolar Amino Acids.” Science, 262: 1680-1685.
To Learn More:
Rebecca Kranz with Andrea Gwosdow, Ph.D. Gwosdow Associates
A note about Wikipedia from Wikipedia: "Wikipedia cannot guarantee the validity of the information found here. The content of any given article may recently have been changed, vandalized or altered by someone whose opinion does not correspond with the state of knowledge in the relevant fields. Note that most other encyclopedias and reference works also have similar disclaimers." http://en.wikipedia.org/wiki/Wikipedia:General_disclaimer.
HOME | ABOUT | ARCHIVES | TEACHERS | LINKS | CONTACT
All content on this site is © Massachusetts
Society for Medical Research or others. Please read our copyright
statement — it is important. |
|
|

Dr. Michael Hecht

Frick Chemistry Laboratory, Princeton University
http://www.princeton.edu/main/news/multimedia/
Sign Up for our Monthly Announcement!
...or  subscribe to all of our stories! subscribe to all of our stories!

What A Year! is a project of the Massachusetts
Society for Medical Research.
|
|

